New approach to seeing inside cells could speed up understanding of molecular machines in their natural context
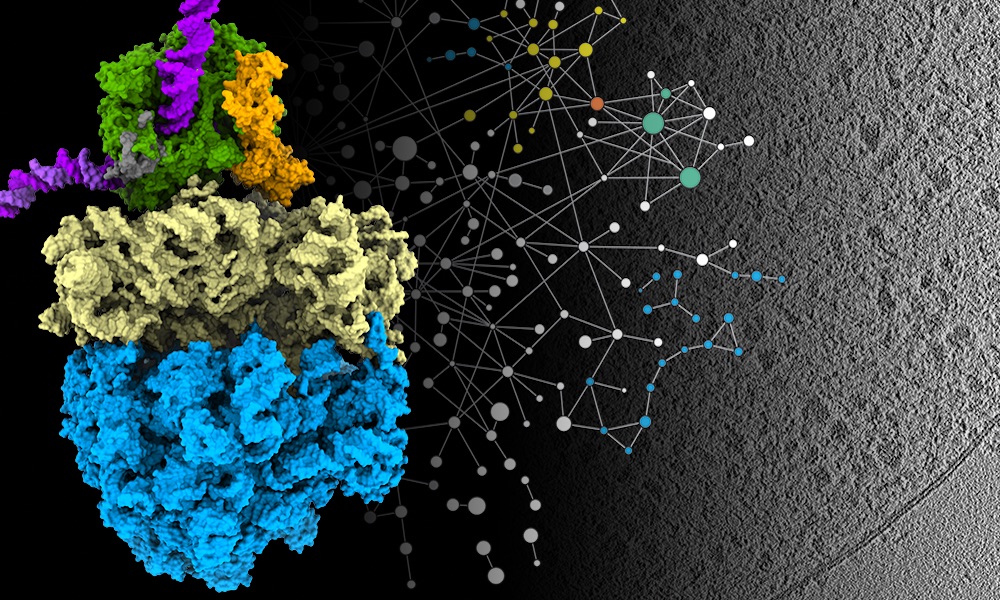
In a paper published this week in the journal Science, researchers in EMBL’s Mahamid group and Juri Rappsilber’s lab at Technische Universität Berlin report how they combined three methods to get high-resolution imagery of parts of the cell that had previously only been studied when removed from the cell and observed in isolation.
One way scientists can study the structure of biological molecules is with a technique known as cryo-electron microscopy. This allows researchers to glimpse structures in high resolution, but often involves creating highly purified samples that are taken out of their natural context of the cell. However, technical advances in electron microscopes, detectors, and computational analysis have advanced cryo-electron tomography – something like a CT scan for cells – to visualise these structures directly inside cells. EMBL group leader Julia Mahamid and her team decided to use cryo-electron tomography to see some of the cell’s key molecular machinery at work inside a bacterial cell.
Two key processes for molecular biologists are transcription and translation – the processes that all cells use to convert information from DNA to proteins. Transcription is carried out by a specialised protein complex called RNA polymerase, which binds to DNA and goes along the DNA strand, using it as a template to create a set of messenger molecules. These messages then go to the cell’s protein-making machines, called ribosomes, which translate them into proteins. In cells that have a nucleus, these processes happen in different locations, with transcription taking place inside the nucleus and translation happening outside it. However, scientists have hypothesised for decades that in bacterial cells – which don’t have a membrane-enclosed nucleus – the two processes are coupled together, and the RNA polymerase and ribosomes can come into physical contact. Julia’s group has now been able to show that this is the case. And they learned that the molecular activity in its natural context inside cells is different from what had been observed in previous studies outside the cell.
The research team combined cryo-electron tomography with cross-linking mass spectrometry and computer modelling to refine their findings, producing the highest-resolution images ever obtained of Mycoplasma pneumoniae – a bacterium that causes a mild form of pneumonia. They studied Mycoplasma in three different scenarios: as it is treated with two different antibiotics and also when it is left untreated.
“We started this project out of curiosity to see how far we could go,” Julia says. “For us, this paper is just the beginning – a proof of principle. Out of this, we have now initiated a number of different projects involving other molecular machines and antibiotics, and with quite a few groups at EMBL interested in applying this approach in their research.”
Julia’s group credits the EMBL environment for helping them achieve these results. Ten years ago, EMBL researchers established Mycoplasma pneumoniae as a model for systems biologists. Additionally, she says EMBL’s Cryo-Electron Microscopy Service Platform is the premier platform of this type worldwide.
“We couldn’t have produced this quality of research within just three years of starting a lab without the data and support we have at EMBL,” Julia says. “This isn’t an incremental advance, but rather a jump. Cryo-electron microscopy revolutionised structural biology a few years ago. Our new approach is likely to contribute to a second revolution where you can study structures directly while still inside cells.”
Source Article
F. O’Reilly, L. Xue, et al. Science, published online 30 July 2020. DOI: 10.1126/science.abb3758
This article was first published on EMBL News
