EMBL-EBI’s MGnify data resource helps researchers find enzymes for novel applications
by Oana Stroe
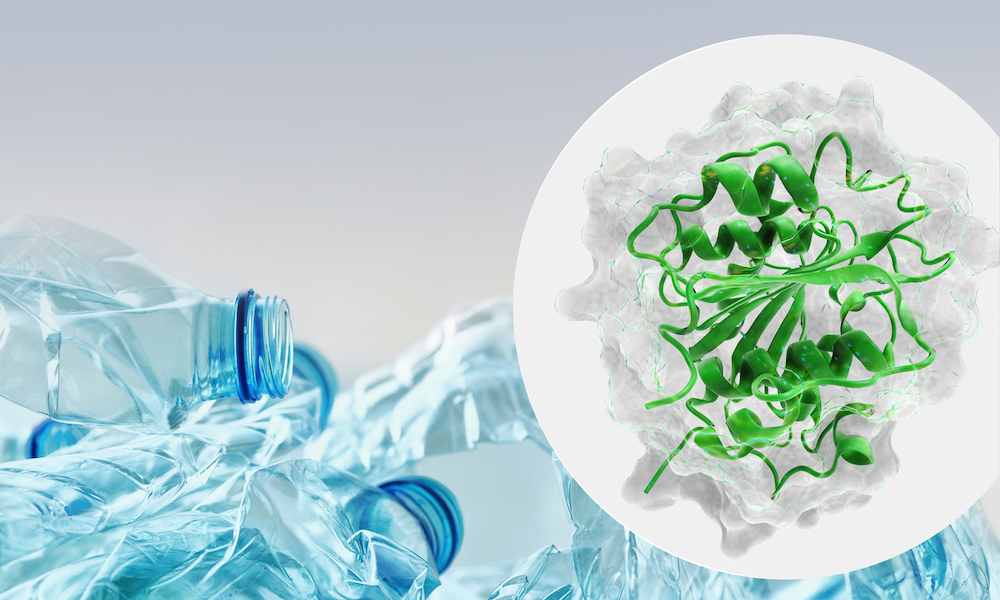
The number of new and poorly characterised proteins available in metagenomic databases, such as EMBL-EBI’s MGnify, is in the billions and growing rapidly as new data from different environments is assembled and analysed.
This represents a treasure trove for the discovery of novel enzymes with exciting properties. However, there is a need for better tools to be able to effectively mine these databases and find enzymes of interest. Thanks to new funding, EMBL-EBI researchers and their collaborators will now be able to turn their considerable expertise towards discovering a special class of enzymes – ones that can degrade plastic also known as ‘plastizymes’.
Platizymes: plastic-degrading enzymes
Plastizymes could be used to reduce the pollution caused by plastic products. Plastic pollution is a fundamental environmental challenge, an enormous waste of resources, and has the potential to become a major world health issue through the ingestion of micro-plastics.
However, there are currently very few known natural plastizymes, and these are relatively inefficient and do not degrade all types of plastic pollutants.
Computational biology meets protein engineering
A new project, funded by UK Research and Innovation (UKRI) aims to address these limitations by employing a combination of computational and protein engineering approaches to discover new plastizymes and improve their catalytic ability.
The team will employ a number of cutting-edge technologies:
- machine learning tools to mine a massive metagenomic database (MGnify)
- ultrahigh throughput directed evolution in microfluidic devices
- fragment-based structural approaches
EMBL-EBI’s MGnify data resource helps researchers find enzymes for novel applications.
This will allow them to simultaneously derive generic pipelines for the discovery and directed evolution of novel enzymes, while exploiting these pipelines to produce improved plastizymes.
“We already know some enzymes that can degrade certain types of plastics, but we want to find better versions, combining data, protein structures and machine learning techniques to determine what makes a good plastic degrading enzyme,” said Rob Finn, Team Leader at EMBL-EBI. “We want to generalise this approach to then tackle different types of plastic that are harder to break down.”
In the longer term, these novel plastizymes could contribute towards the UK’s net-zero ambition by increasing our capacity to recycle plastic waste.
A collaborative project
This project is a collaboration between four investigators based at EMBL-EBI, University College London, and University of Cambridge. The funding is part of the BBSRC’s strategic Longer and Larger (sLoLa) grants programme, which aims to catalyse ground-breaking discoveries that may have far-reaching implications for agriculture, health, biotechnology, and the green economy.
This research also exists as part of EMBL’s Planetary Biology transversal theme, a collaborative effort across all of EMBL and beyond to shed light on the molecular underpinnings and impacts of the pressing societal issues that affect our planet. This includes plastic pollution, CO2 accumulation, antimicrobial resistance, environmental consequences of pesticides, and biodiversity decline, and aims to provide novel solutions for improving planetary health.
Read the full press release on the UK Research and Innovation (UKRI) website.
Related links
This article was first published on EMBL News
