by Vicky Hatch
New software enables scientists to visualise RNA secondary structures using the world’s largest RNA structure dataset
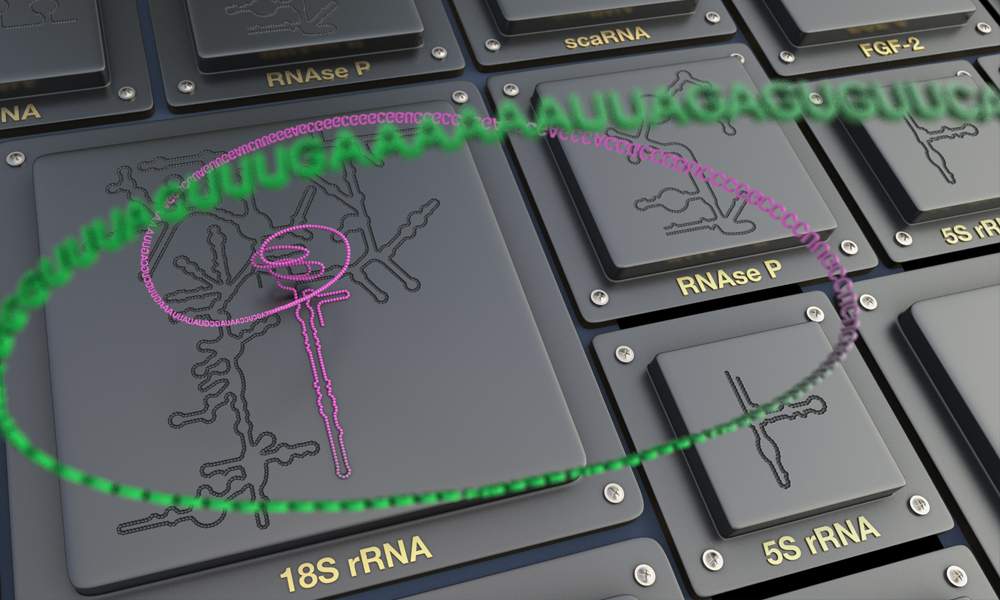
Researchers from EMBL’s European Bioinformatics Institute (EMBL-EBI), Georgia Institute of Technology, Charles University, the National Center for Biotechnology Information, the University of California Santa Cruz, and the University of Texas at Austin have developed R2DT (RNA 2D Templates), a new user-friendly piece of software for visualising 2D RNA structures.
RNA molecules can be large and complex, and their distinct structure is often crucial for their biological function. A structural hierarchy exists in the architecture of RNA molecules. First, there is the RNA sequence – the primary structure. The linear RNA molecule can fold to give the RNA its secondary, 2D structure. Within the secondary structure, interactions occur that cause the RNA molecule to adopt a 3D structure.
R2DT provides a new method for visualising 2D RNA structures using a comprehensive framework that extends across many types of RNA, from very small RNA molecules like transfer RNAs to large ribosomal RNAs. Based on a library of 3,632 experimentally validated RNA structures, referred to as templates, R2DT provides consistent and recognisable RNA layouts, openly available to all researchers.
Consistent, reproducible RNA structures
“Researchers are used to seeing RNA structures in a standard orientation which is widely accepted,” says David Hoksza, Assistant Professor at Charles University. “For larger RNAs such as ribosomal RNA this can be tricky, and to tackle this we developed the template-based drawing method implemented within R2DT. Basically, this method takes a template RNA structure and layout and compares this to a new RNA structure of interest to find commonalities between the two structures.”
R2DT has been extensively tested on over 13 million RNA sequences from the RNAcentral resource, to ensure that the 2D RNA structures it generates are consistent and reproducible. The R2DT framework can also be easily updated and extended with new RNA structures including newly identified viral RNAs. The software is freely available through GitHub and RNAcentral.
“It’s important for researchers to visualise RNA structures in a layout that they will be familiar with,” says Anton I. Petrov, Project Leader at EMBL-EBI. “For decades, researchers have been visualising RNA structures in a way that provides clear meaning to the various parts of the diagrams. This is what we can now produce automatically at a massive scale using R2DT.”
3D information in a 2D structure
The interactions within an RNA structure can be represented in a 2D diagram, used by scientists to investigate the function of RNAs. These 2D diagrams are preferred by researchers as they’re far more accessible and can present a broader variety of information than the corresponding 3D structures. R2DT uses experimentally determined 3D structures and feeds this information into the 2D diagrams to improve the traditional 2D RNA layouts.
“This software allows us to map complex information from 3D RNA structures into a 2D structure which can be visualised much more easily,” says Anton S. Petrov, Co-Investigator at Georgia Institute of Technology. “You could say that these secondary structures are similar to the information we receive from Google Maps; a flat projection of 3D structures but retaining all the vital information.”
Help from the public
The Rfam resource contains data on different RNA families, which are made up of individual RNA molecules with similar structures and sequences. To help explain this idea to a lay audience, Anton I. Petrov and colleagues developed an award-winning public engagement activity called Genome Explorers. This activity gets participants to sort different RNAs by their shapes – their secondary structures generated by R2DT – and put those that look alike into groups.
Watching people sort the RNA structures in this way made it clear to the researchers that the structures needed to be orientated in the same way to allow participants to compare them easily. This gave them several ideas for improving the R2DT algorithm to ensure that RNA structures are generated in the same orientation. They have now deployed this new feature within R2DT, RNAcentral, and Rfam.
Find out more
Learn more about the Genome Explorers activity
This post was originally published on EMBL-EBI News
