by Oana Stroe
The river Cam, which flows through the city of Cambridge, UK, is a hotspot for swimmers, rowers, and kayakers, many of whom develop waterborne infections caused by the bacteria found in the river.

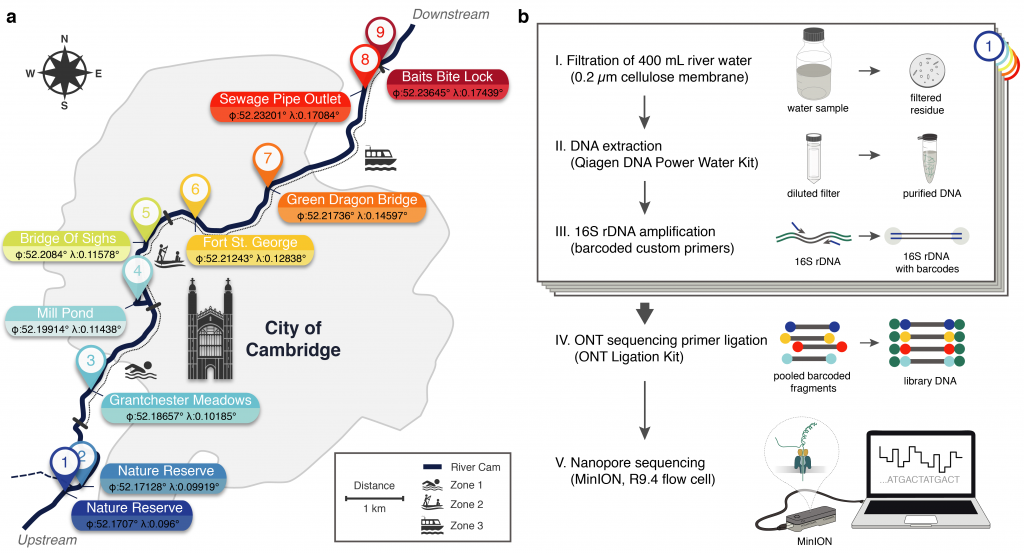
Researchers from EMBL’s European Bioinformatics Institute (EMBL-EBI) and the University of Cambridge have shown that portable DNA sequencing can easily monitor bacterial species in a river ecosystem. They used a smartphone-sized DNA sequencing device to monitor water samples taken from nine locations on the river Cam. Alongside many harmless bacteria, they also found several pathogenic species.
Harmful or not?
The researchers found that potentially harmful and wastewater-related bacteria strongly increased downstream of the most urbanised river sections. One example is Pseudomonas aeruginosa, a bacterium that can cause disease in plants and animals, including humans, varying from pneumonia to sepsis syndromes.
The study, called PuntSeq, also identified the well-known Leptospira bacterium, which can cause the life-threatening disease leptospirosis. However, the Leptospira species found on this occasion were most likely not pathogenic. The full results of the research have been published in the journal eLife.
Quick and easy monitoring
“We set out with the aim of characterising the diversity of microbes in the river,” explains Lara Urban, formerly a PhD student at EMBL-EBI and now a research fellow at the University of Otago, New Zealand. “We also wanted to see how viable it was to use portable DNA sequencers to monitor bacteria species in rivers. With the river Cam on our doorstep, it wasn’t long before we were out collecting samples.”
Traditional microbial tests focus on the isolation of specific bacterial organisms in a laboratory, which is time consuming and reliant on specialised infrastructure and staff. By contrast, the method used in this study – called environmental metagenomics – can identify the DNA of the many bacteria present in a sample of river water. In recent years, devices for mobile DNA analysis have made environmental metagenomics more accessible, allowing real-time sequencing.
A useful tool for public health
“In our study, we report a simple, cost-effective workflow to assess and monitor microbial freshwater ecosystems with targeted nanopore DNA sequencing,” says Maximillian Stammnitz, formerly a PhD student in Cambridge, now a postdoc at the Centre for Genomic Regulation (CRG) in Barcelona. For this study, the researchers successfully quantified the proportions of hundreds of different bacteria at nine sites along the river. For most locations, the researchers collected several samples throughout the spring and summer months.
“This study design has allowed us to provide real transparency on important local questions of public health, with a very exciting new technology,” says Andre Holzer, a PhD student at the University of Cambridge.
“We believe this method could be useful for public health, local authorities, and researchers studying waterborne diseases around the world,” Urban adds.
This post was originally published on EMBL-EBI News.
