by Fabian Oswald
Stephen Cusack, Head of EMBL Grenoble, discusses how the influenza virus infects cells, and shares his most recent discoveries.
The infectious disease commonly known as flu is caused by the influenza virus. It spreads around the world in seasonal outbreaks, causing millions of infections and hundreds of thousands of deaths each year. Stephen Cusack, Head of EMBL Grenoble, has been studying different aspects of the influenza virus for 30 years. He recently published a paper describing the function of influenza polymerase – a key enzyme of the virus – in unprecedented detail. Here, he describes how the influenza virus functions, the technologies that are helping researchers to understand it, and why it’s necessary to constantly develop new drugs.
Tell us about your research on the influenza virus.
We study how the influenza virus proliferates in an infected cell; in particular, how the genome of the virus is copied by the viral polymerase. Most cellular organisms and some viruses have a genome made of DNA. The influenza virus is an RNA virus, which means that its genome is made of RNA – a molecule similar but chemically distinct to DNA. The viral genome serves two purposes. Firstly it is transcribed: this is the process by which it is used to make what’s called messenger RNA, or mRNA. This mRNA carries instructions to the protein-making machinery of the host cell to make viral proteins. Secondly, the viral genome has to be replicated, which means that exact copies of it are made. These are packaged, together with the newly synthesised viral proteins, into the next generation of viruses, which bud at the cell surface.
What happens when the virus infects a cell?
The virus attaches itself to the cell surface. It then enters the cell through a process called endocytosis. This means that the virus becomes surrounded by part of the cell membrane, forming a kind of bubble that detaches from the surface and is pulled into the cell. The RNA genome is released from inside the virus and transported into the nucleus of the infected cell. The viral polymerase is brought into the nucleus already tightly bound to the genome in such a way that it can get started straight away, first with transcription and then – a few hours later, after additional viral proteins have been made – with replication of the genome.
And what is the focus of your recent research?
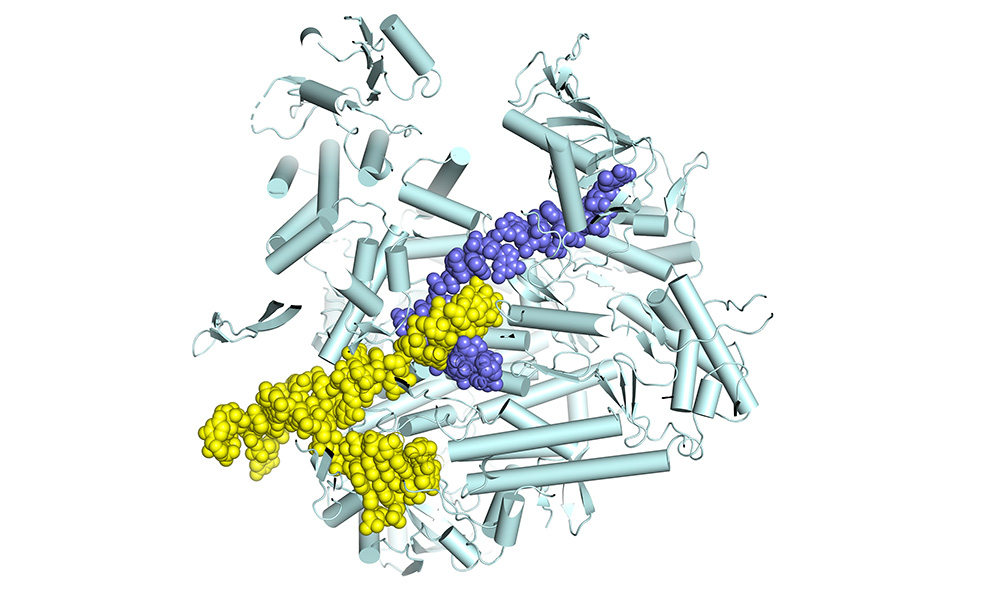
The polymerase is a molecular machine, moving along the RNA genome as it copies it. We want to find out what all the moving parts are and thus understand how the machine works. Then we will be in a good position to develop antiviral drugs that stop the polymerase in its tracks. To do all that, we use structural biology to determine the atomic structure of the polymerase. However, you need not just one structure, but many snapshots of the machine in action. That’s what we’ve been doing, step by step, for the past twelve years. We started out by determining the structure of fragments of the polymerase. This was in itself interesting, as some of these fragments are targets against which highly potent anti-influenza drugs have been developed. Then, in 2014, we were able to determine by X-ray crystallography the structure of the complete polymerase, which is a really big and complex molecule, made of three separate protein chains folded together. At that time, it was still unclear how it really worked, because the first structures didn’t show the polymerase in an active state – when it’s actually copying the genome. Since then, we’ve spent a lot of time figuring out how to determine structures of the functioning enzyme. That has only really become possible since the so-called cryo-electron microscopy resolution revolution.
Why was this essential?
Cryo-electron microscopy, or cryo-EM, is a technique that now allows us to get structures of protein complexes at near atomic level. This has only been possible in the last five or so years, thanks to spectacular technical advances. Prior to that, we used X-ray crystallography, a powerful technique that can also give high resolution, but to do it you need to grow crystals of your sample. In the crystal, all the molecules are constrained to be in the same state, and that’s not always possible when the molecule is very flexible. With cryo-EM, the sample in solution is applied to grids and then rapidly frozen, trapping the polymerase particles, each in its particular state, in a thin layer of ice. The grids are then placed in the electron microscope and many images captured of different parts of the grid where there are particles. We use sophisticated image processing software to process the data to obtain a high-resolution structure. That’s what we’ve been doing for the past three years, with the support of the Cryo-EM Service Platform at EMBL Heidelberg, and it’s worked beyond all expectations. A few years ago, we would never have believed this was possible.
Could you explain your most recent publication?
We’ve now managed to understand how the influenza polymerase carries out the complete transcription process. We set up a transcription reaction in the test tube in a way that meant it would stall at different points in the process. For each stalled reaction, we determined the structure of the polymerase bound to the genomic RNA and to the partially made product mRNA. In this way, we obtained a series of snapshots of eight successive stages, from the beginning to the end of transcription, resembling a molecular movie. This has never been done before in such detail for a large viral polymerase. We were able to reveal some unique features of influenza polymerase for the first time. For instance, we show how the polymerase copies the genome, but never lets go of the genome ends, which enables rapid recycling for the next round of transcription. This is really breakthrough research, and has now been published in Cell. It is mainly the work of PhD student Joanna Wandzik and EMBL Interdisciplinary Postdoc Tomas Kouba. Biochemical Technician Petra Drncova made a very instructive animation of the complete transcription cycle, which can be viewed below.
What avenues does this open for your future research?
There are a few things we want to do next. First, we want to apply this method to the other activity that the polymerase has, when it replicates its genome to be packaged into new viruses. This is more complicated, as there may be two polymerases involved. Then, we want to move towards the more physiological aspects. Until now, we’ve used influenza polymerase made in the lab and working in a test tube. This enables you to understand the basic mechanisms. But in reality, the RNA genome is never naked but at all times packaged together with the polymerase and many copies of the viral nucleoprotein in the so-called viral ribonucleoprotein particle (vRNP). The processes of replication and transcription actually take place in the vRNP context. We want to understand how that happens. The third step will be to go really into the cell and look directly at the polymerase working in the nucleus, as certain host proteins are crucial for transcription and replication as well. This is very challenging, but is becoming possible using the developing technique of cellular cryo-electron tomography. There’s a lot of exciting work on that going on at EMBL Heidelberg, particularly in the group of Julia Mahamid, who’s pioneering this technique.
Another important project we will focus on is developing new drugs that help to treat influenza. The virus evolves quickly, thanks to errors that the polymerase makes in copying the genome, and can develop resistance to drugs, so there’s always a need for the next generation of drugs. All the studies we’ve done have opened up opportunities for developing new ways of targeting the polymerase as part of an antiviral drug development program.
Can your methods for studying influenza be adapted to other types of viruses?
Yes, very much so. We’re currently extending our studies from influenza to other related viruses that have a similar kind of polymerase, but infect the cell in very different ways. Together with the Virology Department at the Bernhard Nocht Institute for Tropical Medicine in Hamburg, we’re studying the Lassa virus, which sporadically produces local epidemics of Lassa fever in Western Africa, and is very dangerous. More generally, all RNA viruses have an RNA-dependent RNA polymerase, and they’re all evolutionarily related. The structure of the core of the enzyme is always the same, and in principle the methods we used can be applied to any of them. Many human pathogens are RNA viruses, including the common cold virus, Ebola, HIV, and SARS-CoV-2, which is causing the current COVID-19 pandemic. SARS-CoV-2 is not in the same family as the influenza virus, but it also has an RNA polymerase that fundamentally works in the same way, because it has to copy its RNA genome. Many people are trying to gain structural insights that will help target the SARS-CoV-2 polymerase for drug development. Developing a completely new drug will take a long time. However, some polymerase inhibitors already developed and tested against influenza or Ebola viruses seem to be active against SARS-CoV-2, and this brings some hope that treatments for COVID-19 may become available soon.
An animation showing the transcription cycle of the influenza virus. The RNA (black) is pulled through the viral polymerase while being read and transcribed into a messenger RNA (blue). VIDEO: Petra Drncova/ EMBL
This articel was first published on EMBL News
